Research
Histone Lysine Demethylases – PHF8 (KDM7B)
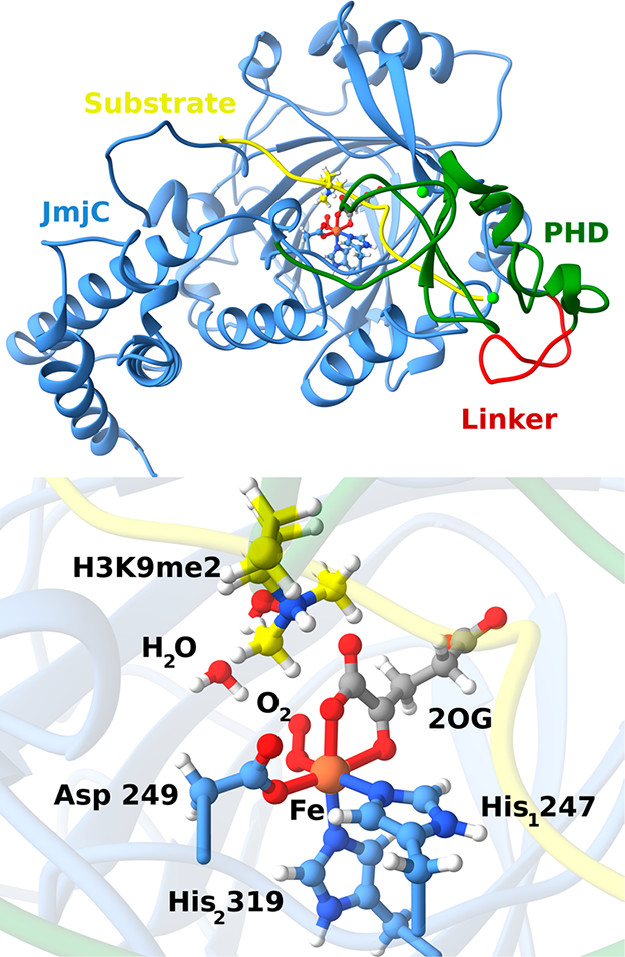
The histone lysine demethylases (KDM2-7 subfamilies in humans) are Jumonji C (JmjC) domain-containing enzymes. The JmjC proteins are Fe (II) and 2-oxoglutarate- (2OG-) dependent dioxygenases that couple substrate oxidation to decarboxylation of 2OG to form succinate, CO2, and a ferryl−oxo intermediate. The latter performs hydrogen abstraction from the substrate, followed by hydroxylation and nonenzymatic formation of formaldehyde.
PHF8 (KDM7B) catalyzes the demethylation of the dimethylated or monomethylated lysine 9 residue in histone H3 (H3K9me2/me1), in a manner actively promoted by binding of its N-terminal plant homeodomain (PHD) to H3K4me3.
Adapted from: ACS Catal. 2020, 10, 2, 1195–1209.
Ethylene Forming Enzyme
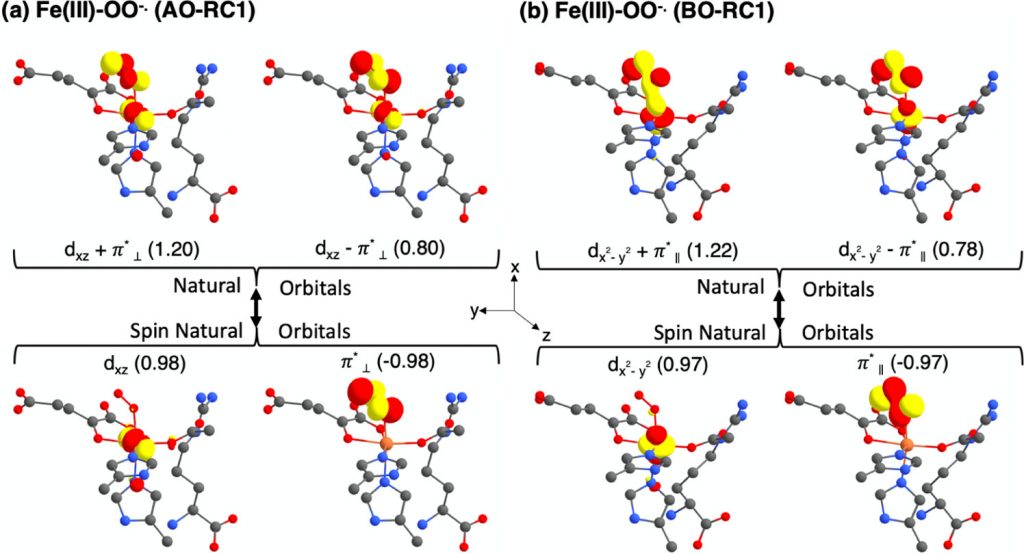
The ethylene-forming enzyme (EFE) is a non-heme Fe(II), 2- oxoglutarate (2OG)-, and L-arginine (L-Arg)-dependent oxygenase that catalyzes two reactions. The less prominent EFE-catalyzed transformation involves oxidative decarboxylation of 2OG, forming succinate and CO2, coupled with C5 hydroxylation of L-Arg, where the hydroxylated L-Arg spontaneously decomposes to guanidine and L-Δ1-pyrroline-5- carboxylate. The major EFE-catalyzed reaction involves the decomposition of 2OG to ethylene plus three molecules of CO2/bicarbonate. EFE’s hydroxylation reaction is typical of many other Fe(II)/2OG oxygenases in this superfamily;3−6 however, the generation of ethylene by cleaving three C−C bonds of 2OG is unique to this enzyme.
Adapted from: ACS Catal. 2021, 11, 3, 1578–1592.
DNA Hydroxylases and Demethylases
Alkylation of DNA bases by endogenous and exogenous sources can cause cytotoxicity and/or cancer-linked mutations. Direct repair of damaged DNA bases occurs via processes involving DNA glycosylases and AlkB type oxygenases. AlkB family oxygenases utilize 2-oxoglutarate (2OG) and Fe(II) to catalyze the demethylation of alkylated DNA bases. DNA demethylases include bacteria AlkB and its human homologues, AlkBH1 to AlkBH8, and FTO. AlkB and AlkBH3 prefer to repair methylation damage in ssDNA rather than dsDNA, whereas AlkBH2 efficiently acts on dsDNA compared to ssDNA. They act on multiple monoalkylated and exocyclic-bridge DNA nucleobases such as m1A, m3C, m1G, m3T, eA, and eC.
Adapted from: ACS Cent. Sci. 2020, 6, 5, 795-814.
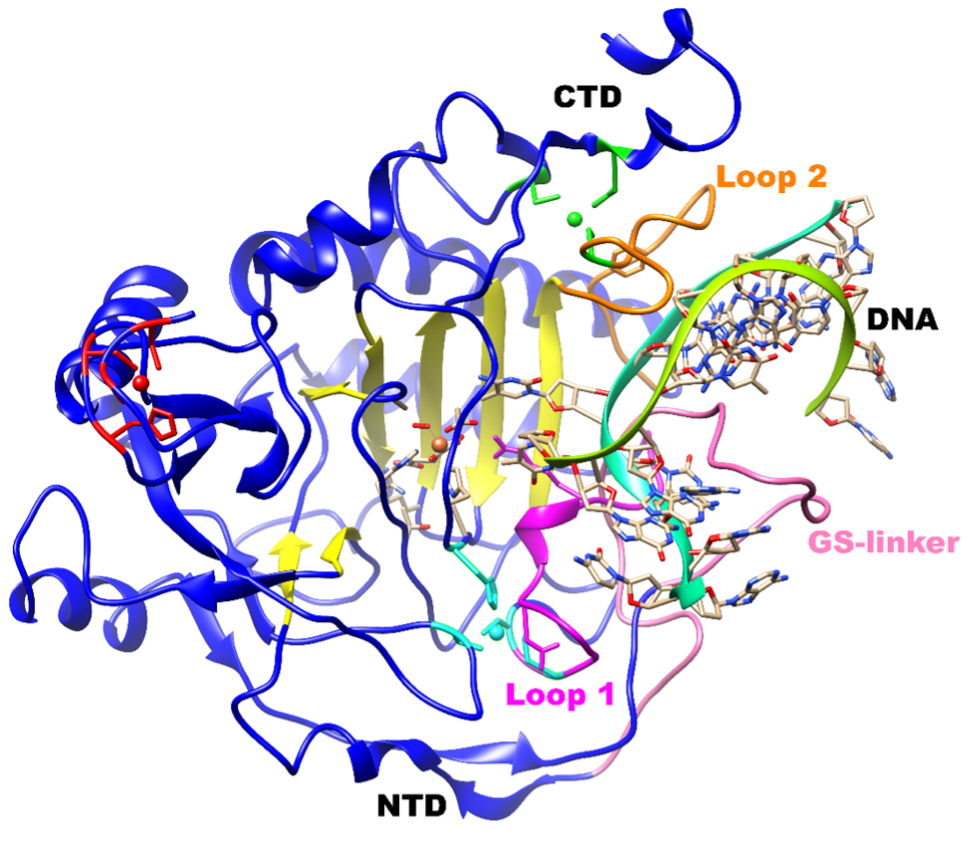
TET proteins, which are also non-heme Fe(II) and 2OG dependent enzymes, are classified as DNA hydroxylases that perform successive oxidation of 5mC to 5hmC, 5fC, and 5caC. TET enzymes contain a conserved double stranded b helix (DSBH) domain, a cysteine-rich domain, and binding sites for Fe(II) and 2OG co-substrate that together form the core catalytic region in the C-terminus.
Adapted from: ACS Catal. 2021, 11, 7, 3877-3890.